Multiple Hypothesis Testing
Multiple Hypothesis Testing
Similar to the setup for single hypothesis testing, we consider data , where . However, here we test multiple null hypotheses: for . Here is the number of hypotheses, and are general subsets of that may overlap. A straightforward example is that is a null hypothesis about the -th coordinate of , and contains parameters with their -th coordinate satisfying the null condition.
Gaussian
Consider Gaussian data with . One example multiple testing problem is to test against .
Genome-wide association studies
Given some disease status variable , we want to study the association of each gene with the disease. Consider with , where is some gene. We test against is independent of .
We ask the following questions.
Questions
- (Global null testing). Is true? For example, when the null hypothesis is , the global null testing asks whether holds.
- Which are not true? We ask this because the effects of the alternative hypotheses are often of greater interest.
Similar to Evaluating a Test, different questions (tasks) lead to different evaluation metrics. Let the output of a multiple hypothesis test be the rejection set . Let be the test statistic for , and the rejection region for be . That is, . We consider the following two error metrics:
Family-wise error rate (FWER)
We want to return an such that . Equivalently, we want to find the critical values such that .
False discovery rate (FDR)
We want to return an such that at most an -fraction of the rejected hypotheses are null. Equivalently, we want to control . The elements in are called discoveries.
FWER obeys the Neyman-Pearson paradigm which controls the Type-I error, which can be too conservative in high-dimensional settings. Instead, FDR restricts the scope of discoveries and consider a relative error rate
A key question in designing a multiple testing algorithm is how to combine the results of individual hypothesis tests to produce a coherent output. p-values serve as a convenient object to work with for this purpose. We denote as the p-value for , i.e., for all and . ^[Note that by definition p-values are super-uniform; but sometimes we assume they are exactly uniform to obtain tight results.] The following figure plots the sorted p-values for different signals. Specifically, when the null hypothesis is true, the p-values are uniformly distributed (no interesting signal); when the sorted p-values deviate significantly from the line , it presents a clear signal. However, this signal does not directly translate into that all p-values below a certain threshold are significant. Because even when all nulls are true, due to the high dimension, some of their p-values will be small by chance. Thus, the observed signal suggests only a systematic departure from the null hypothesis rather than significance for each individual p-value.

A multiple testing algorithm using p-values is of the form
One simple algorithm of this kind is the Bonferroni algorithm.
Bonferroni Algorithm
Let be the family-wise error rate (FWER). Let be the p-values of individual tests. The Bonferroni algorithm returns
FWER control for Bonferroni
The Bonferroni algorithm controls the FWER at level .
Proof
By definition, the FWER is
where . Then, by the union bound,
We remark that the Bonferroni algorithm works for dependent tests. Nonetheless, the following example on independent Gaussian helps us understand the algorithm.
Gaussian
Consider data , null hypotheses , and one-sided p-values . Then, the Bonferroni algorithm rejects if , which is equivalent to . See ^fig-g for a visualization of how the Bonferroni algorithm controls the cumulative tail probability.

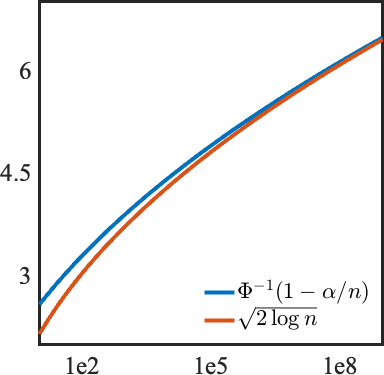
The following proposition gives an approximation of for any .
Max-Central Limit Theorem (Fisher–Tippett–Gnedenko) ^prop-max
^fig-a gives an illustration of how approximates when .
Sparsity Connection
The previous proposition already connects the threshold of the Bonferroni algorithm to the max statistic, which is good for detecting sparse signals. The following propositions further formalize the connection between the Bonferroni algorithm and sparsity, suggesting that the Bonferroni algorithm is good at detecting sparse signals and dealing with sparse alternatives.
Proposition
If with and for , then the Bonferroni algorithm has power
Proof
First, by the definition of the Bonferroni algorithm, we have
where . Then, by Proposition ^prop-max, we get
Letting gives
The following proposition can be obtained by a similar argument.
Proposition
If with and for , then the Bonferroni algorithm has power approaching 0 as .
Benjamini-Hochberg Algorithm
Let be sorted p-values. The Benjamini-Hochberg (BH) algorithm returns
That is, denoting , we reject nulls with the smallest p-values.
Note that
That is, unlike Bonferroni, BH does not reject nulls under a function graph. Instead, it first determines , and then reject nulls before on the line of sorted p-values. See ^fig-bh for an illustration: even before there are some p-values above the line , they get rejected because is the last index below the line. Also note that without the bound’s dependence on , the BH algorithm reduces to the Bonferroni algorithm.

FDR control for BH
For independent p-values, the BH algorithm satisfies . The first equality holds when the p-values are uniformly distributed for true nulls.
Proof
We first define the confusion matrix:
# Accepted Rejected True False And we denote as the index set of true nulls; is the rejection set as before. With a slight abuse of notation, we use and to also denote the number of true nulls and the number of rejections, respectively. Then, , , , and .
Define the indicator . By definition, we have
Without loss of generality, we assume the p-values are already sorted. Then, observe that if , there exists such that . Therefore, if we consider a virtual instance where and other p-values are unchanged, BH will return the same rejection set for this instance.^[For this virtual instance, p-values may not be sorted anymore. However, p-values that are smaller than in the original instance will stay below ]. We denote as the rejection set after setting to 0. Note that this virtual instance only depends on . Therefore,
where the first equation is because if , setting will not change the rejection set as we argued; the second equation uses the tower property; the third equation uses the fact that is independent of , and the indicator specifies that ; the fourth inequality uses the fact that is super-uniform, and the equality holds when it’s uniform.
Connection Between Different Metrics
Using the confusion matrix, we can express the metrics as
Since , we have , and thus . For a single hypothesis test, or if we consider the global null, where we either reject all nulls or none, we have . In this case, .